We are a structural biology and drug discovery laboratory. Our research focuses on developing small-molecule chemical drugs for anticancer and antiviral therapies. Our primary targets include key proteins in the ubiquitination system and kinase families, such as deubiquitinating enzymes (DUBs) and the pseudokinase RNase L. Utilizing structure-based drug design and fragment-based drug discovery approaches, we aim to develop highly specific and potent lead compounds. Our goal is to create drugs with independent intellectual property rights in China.
We also conduct signaling and cell biology research in the fields of the ubiquitin-proteasome system and kinase families, with a focus on pathways closely associated with cancer development. We seek to elucidate the regulatory roles of key proteins in cancer progression and identify novel targets for anticancer drug development.
In the kinase research area, we have extensive experience and a unique systematic research approach in the pseudokinase RNase L project.
Our research projects directly bridge fundamental science and drug discovery, striving to integrate chemical biology, structural biology, and cell biology effectively in drug development. We welcome students, postdoctoral fellows, and research assistants from chemistry, chemical engineering, biology, pharmacy, medicine, and related disciplines to join our team!
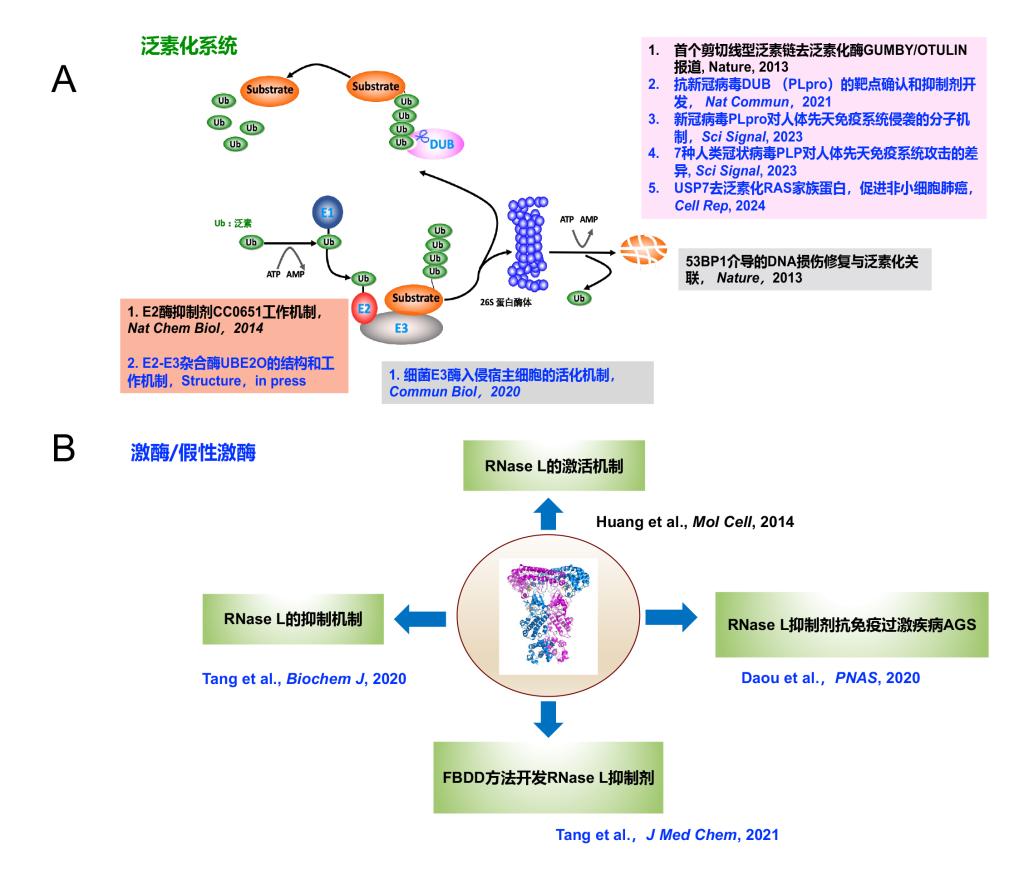
The blue text in the figure above indicates achievements where the first author’s affiliation is Peking University Shenzhen Graduate School.
Published Articles (#: co-first author; *: co-corresponding author)
1) Huang H*#,Zhu W#, Fu Z#, Huang B#, Xiong Y#, Cao D, Ye Y, Chang Q, Li W, Li L, Zhou H, Niu X, Zhang W.Structural insights into the biochemical mechanism of the E2/E3 hybrid enzyme UBE2O,Structure, 2024, in press
2) Huang B#, Cao D#, Yuan X, Xiong Y, Chen B, Wang Y, Niu X, Tian R,Huang H*. USP7 Deubiquitinates KRAS and Promotes Non-Small Cell Lung Cancer,Cell Rep, 2024, 43(11):114917.
3) Liu M#, Li J#, Liu W#, Yang Y#, Zhang M, Ye Y, Zhu W, Zhou C, Zhai H, Xu Z*, Zhang G*,Huang H*. The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors,Angew Chem Int Ed, 2023, 9;62(41):e202309657
4) Cao D#, Duan L#, Huang B#, Xiong Y, Zhang G*,Huang H*, The SARS-CoV-2 papain-like protease suppresses type I interferon responses by deubiquitinating STING,Sci. Signal., 2023,16(783):eadd0082.
5) Xiong Y#, Huang B#, Yang Y, Fu X, Fu Z, Xu H, Liu M, Cao D, Zhang M, Yang H, Niu X, Yu C,Huang H, Papain-like proteases from seven human-infecting coronaviruses differ in substrate selectivity and innate immune suppression,Sci. Signal., 2023,16(783):eade1985.
6) Fu Z#, Huang B#, Tang J#, Liu S#, Liu M, Ye Y, Liu Z, Xiong Y, Zhu W, Cao D, Li J, Niu X, Zhou H, Zhao YJ, Zhang G*,Huang H*,The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery,Nat. Commun.2021;12(1):488.
7) Ye Y#, Xiong Y#,Huang H., Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8,Commun Biol.2020;3(1):752.
8) Daou S#, Talukdar M#, Tang J#, Dong B, Banerjee S, Li Y, Duffy NM, Ogunjimi AA, Gaughan C, Jha BK, Gish G, Tavernier N, Mao D, Weiss SR,Huang H*, Silverman RH*, Sicheri F*, A phenolic small molecule inhibitor of RNase L prevents cell death from ADAR1 deficiency,Proc Natl Acad Sci U S A. 2020;117(40):24802-24812.
9) Huang H, Zeqiraj E, Dong B, Jha BK, Duffy NM, Orlicky S, Thevakumaran N, Talukdar M, Pillon MC, Ceccarelli DF, Wan LC, Juang YC, Mao DY, Gaughan C, Brinton MA, Perelygin AA, Kourinov I, Guarné A, Silverman RH, Sicheri F, Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon induced antiviral activity. Mol. Cell.2014; 53, 221-34
10) Huang H*, Ceccarelli DF*, Orlicky S*, St-Cyr DJ, Ziemba A, Garg P, Plamondon S, Auer M, Sidhu S, Marinier A, Kleiger G, Tyers M, Sicheri F, E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin.Nat. Chem. Biol.2014 10, 156-63





