The Yuqian Chen lab from the National Key Laboratory of Chemical Genomics and the School of Chemical Biology and Biotechnology at Peking University Shenzhen Graduate School, has published a research paper titled "Dual Modality Feature Fused Neural Network Integrating Binding Site Information for Drug Target Affinity Prediction" in the top-tier digital medicine journalnpj Digital Medicine(Impact Factor = 12.4, ranked in the top tier of the Chinese Academy of Sciences). The researchers have achieved a significant breakthrough in the field of drug-target affinity prediction. The team developed an innovative dual-modality neural network model capable of accurately predicting the binding affinity between drugs and target proteins—an advancement expected to significantly accelerate the drug development process.
Accurately predicting binding affinity between drugs and targets is a critical step in drug development. However, traditional experimental methods are time-consuming and costly. Although artificial intelligence technologies have made notable progress in this field, existing computational approaches often struggle to simultaneously leverage both sequence and structural information of drugs and proteins, and they face challenges when handling the scale differences between drug molecules and proteins. To address these key difficulties, the authors designed DMFF-DTA, a dual-modality neural network-based model for drug-target affinity prediction. DMFF-DTA innovatively integrates a sequence-modality feature extraction module and a graph-modality feature extraction module to model both drugs and targets across sequence and graph structural dimensions. Furthermore, to address the size imbalance between drug and protein graphs, the model introduces a binding site contact map construction method based on AlphaFold2.
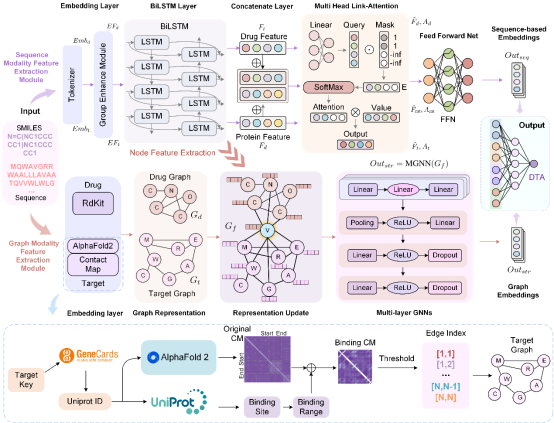
Figure 1: The Framework and Workflow of the DMFF Model
This model effectively integrates multi-dimensional information from both drugs and targets, leading to a significant improvement in prediction accuracy. Experimental results on two standard benchmark datasets (Davis and KIBA) demonstrate that the DMFF-DTA model's predictive performance is notably superior to existing methods. On the Davis dataset, the model reduced the Mean Squared Error (MSE) by 3.6% and increased the Concordance Index (CI) by 0.3%. Similarly, on the KIBA dataset, the model achieved a 3.4% reduction in MSE and a 0.5% improvement in CI. These improvements are all statistically significant (P < 0.05).
Through detailed interpretability analysis, the research team confirmed that the model can accurately identify key regions in drug-target interactions. Statistical analysis revealed that attention values in binding site regions and binding range areas were significantly higher than in other regions (t-test, p < 0.05). Case studies further demonstrated that the model particularly focuses on functional groups within drug molecules (such as O, N, Cl, and F atoms) and benzene ring structures, which is highly consistent with actual binding patterns.
To validate the model's practical value in real-world drug development, the research team conducted a drug repositioning study using pancreatic cancer as a case example. Through systematic analysis, including pathway mapping and evaluation of drug physicochemical properties, potential candidate drugs such as Noscapine were successfully identified. Molecular docking results further verified the reliability of the predictions, demonstrating the model's promising application prospects in practical drug development.
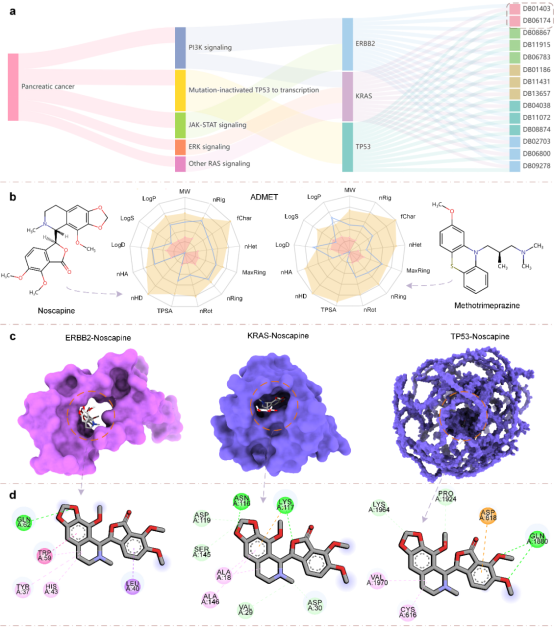
Figure 2: Case analysis results of the DMFF model for pancreatic cancer drug repurposing
In this study, the authors introduced DMFF-DTA, an innovative dual-modality neural network model. It effectively integrates sequence and structural information from both drugs and targets to achieve accurate drug-target affinity prediction. Additionally, a unique AlphaFold2-based binding site graph construction method was developed to effectively address the scale disparity between drug molecules and proteins. Overall, the proposed DMFF-DTA model demonstrates significant advancements in predictive accuracy, computational efficiency, and interpretability through its innovative dual-modality feature fusion strategy and efficient binding site modeling approach. Moreover, its excellent generalization capability and validation through practical case studies make it a powerful tool in the field of drug development, offering a novel solution to accelerate the drug discovery process.
The related findings have been published in the journalnpj Digital Medicineunder the title "Dual modality feature fused neural network integrating binding site information for drug target affinity prediction."Yuqian Chen from theNationalKey Laboratory of Chemical Genomics and the School of Chemical Biology and Biotechnology at Peking University Shenzhen Graduate Schoolis the corresponding author, and graduate student Huaihuai He is the first author.
参考资料
He, H., Chen, G., Tang, Z. et al. Dual modality feature fused neural network integrating binding site information for drug target affinity prediction. npj Digit. Med. 8, 67 (2025).





